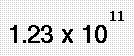What is Molecular Modeling?
Molecular Modeling, is one of the fastest growing fields in science. It may vary from building and visualizing molecules (click image below for a short animation)...
...to performing complex calculations on molecular systems.
Molecular models fall into four basic categories: skeletal or line; stick, ball-and-stick, and space-fillied or CPK.




Some Basic Aspects of Molecular Modeling
Molecular mechanics is one aspect of molecular modelling, as it involves the use of classical mechanics (Newtonian mechanics) to describe the physical basis behind the models. Molecular models typically describe atoms (nucleus and electrons collectively) as point charges with an associated mass. The interactions between neighbouring atoms are described by spring-like interactions (representing chemical bonds) and Van der Waals forces.
Molecular mechanics (MM) is the simplest and fastest way to evaluate molecular systems. For more details see: Introduction to Molecular Mechanics and Molecular Dynamics
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4026342/
Molecular dynamics (MD) is an important computational tool for understanding the physical basis of the structure, the dynamic evolution of the system, and the function of biological macromolecules.
Below, molecular dynamics simulations are performed on a lipid-protein complex. Shown are the backbone structure of the protein, a bound fatty acid molecule within the protein, and a small shell of water surrounding the protein. The time for the simulation was 100 picoseconds. The graphs give information about the motion of the lipid inside the protein.
See: Molecular Dynamics Simulations of Adipocyte Lipid-Binding Protein: Effect of Electrostatics and Acyl Chain Unsaturation for more information.
Below a 380 nanosecond Molecular Dynamics Simulation shows the dynamics of water freezing to ice.
See: M. Matsumoto, S. Saito, and I. Ohmine, Molecular dynamics simulation of the ice nucleation and growth process leading to water freezing, Nature 416, 409-413 (2002); http://dx.doi.org/10.1038/416409a for details.Docking
Molecular docking is one of the most frequently used methods in structure-based drug design, due to its ability to predict the binding-conformation of small molecule ligands to the appropriate target binding site.
Example:

Crixivan (molecule - carbon shown in grey-oxygen-red) is a prescription medicine used to treat the symptoms of HIV Infection. Shown is crixivan bound to the protease enzyme active site. See: How do drugs work
MATHMOL
- Activity 1: Measuring length and distance at the molecular level
- Activity 2: Geometry-of-1-Dimension
- Activity 3: Geometry of 2- Dimensions
- Activity 4: Geometry of 3-Dimensions
- Activity 5: Introduction to Molecular Modeling using Jsmol
- Activity 6: The Geometry of Crystals
- Activity 7: Summary Sheet by Students
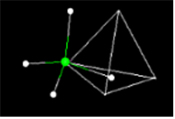
- Activity 8: What is the Geometry of the Methane Molecule
- Activity 9: Geometry of the Crystal Structure of Ice
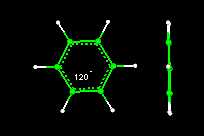
- Activity10:Geometry of the Benzene Molecule
 217K Movie
217K Movie 931K Movie
931K Movie